The Centers for Medicare & Medicaid Services (CMS) regulates all non-research laboratory testing performed on humans in the U.S. through the 1988 Clinical Laboratory Improvement Amendments (or CLIA for short). Many of the tests performed are genetic tests that assess DNA and/or RNA. For years this amounted mostly to PCR and blotting methods. However, more and more clinical labs are employing a myriad of platform technologies to conduct the CLIA lab genetic testing. These include some of the most up-to-date methods such as next generation sequencing (NGS) and digital PCR (dPCR) as well as the vetrans like endpoingt PCR and Southern and northern blotting.
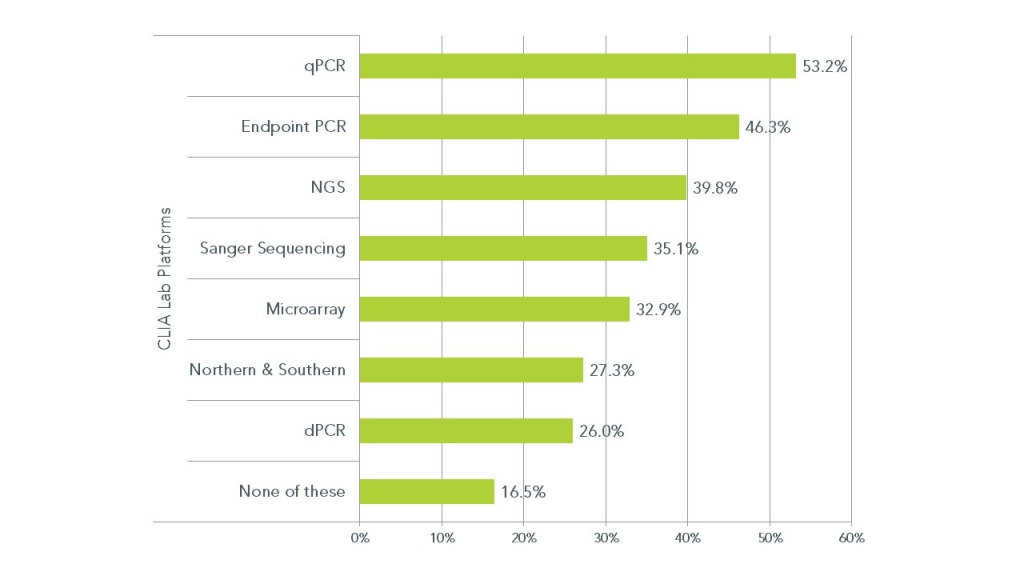
Percepta asked over 230 CLIA laboratories which genetic analysis platform technologies they were currently using. The results are shown in the table below.
PCR, the most commonly deployed CLIA lab genetic testing tool, is present in roughly half of all the 231 labs reporting in this poll. PCR comes in two formats, endpoint PCR and quantitative or realtime PCR (also rtPCR and qPCR). However, NGS is now deployed in roughly 40% of all CLIA labs and has surpassed Sanger sequencing as the most commonly used DNA (or RNA) sequencing method for CLIA lab genetic testing.
Still in use by more than 25% of CLIA labs are older technologies like Southern and northern blotting. But even relatively new technologies such as dPCR are currently deployed by a quarter of all the labs surveyed. This suggests that the older technologies like blotting, Sanger sequencing and PCR are not necessarily being replaced but are supported by the newer technologies like NGS and dPCR in CLIA lab genetic testing.
It is also interesting to note that roughly 17% of all participating labs are using none of these technologies, suggesting that genetic testing may not be on their menu of testing options.


Leave a Reply